Features include:. Fixed length, fixed angle drawing. Automatic alignment of figures.
Detects structures, text, and arrows and places them automatically. Can automatically draw rings and other structures - has all standard amino acids and nucleic acids in built-in library.
Xdrawchem For Mac
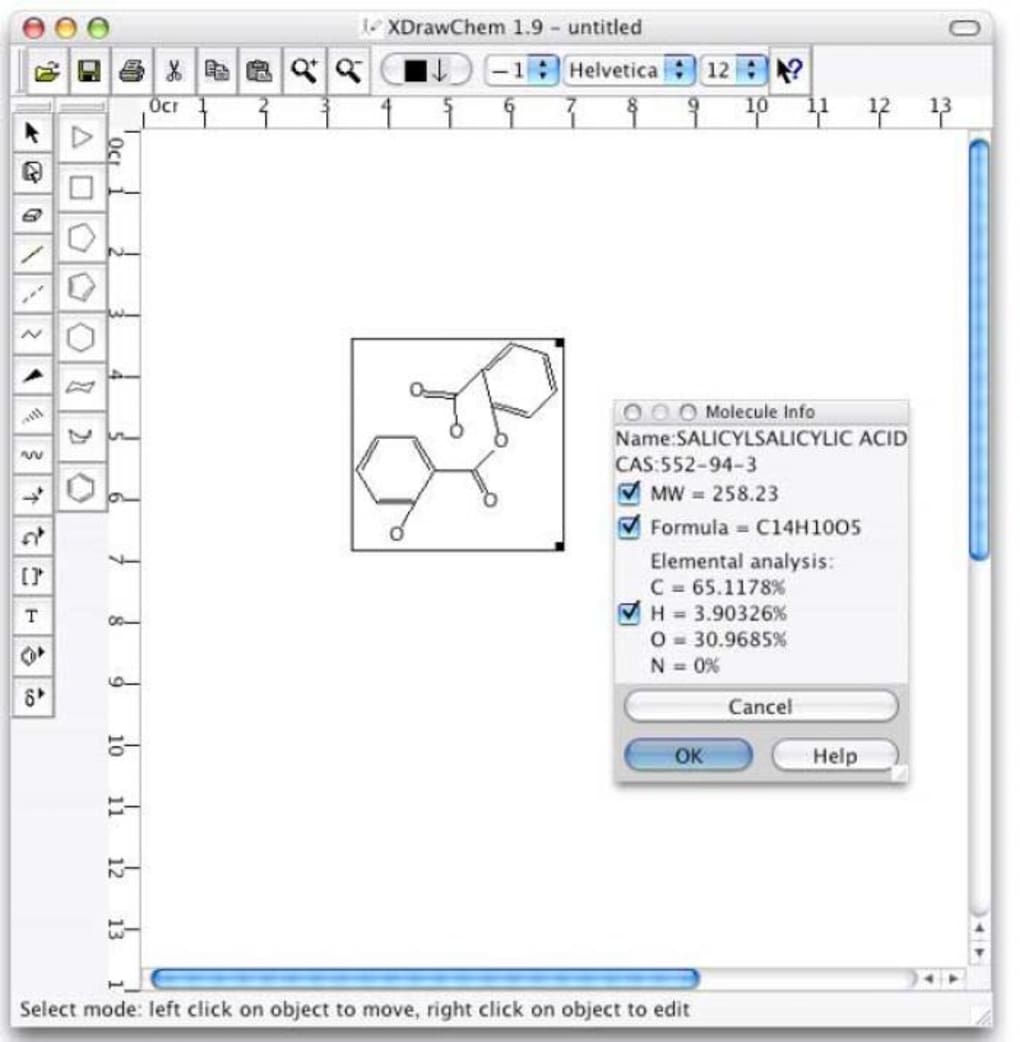
Can retrieve structures from a network database based on CAS number, formula, or name. Can also retrieve information on a molecule in a drawing. The data file (05 August 2005, comma-separated, 60 MB) is freely available. This database was derived from PubChem data as of July 2005. Can draw symbols such as partial charge, radicals, etc.
Xdrawchem For Mac
Can read MDL Molfiles, CML Chemical Markup Language, defined in J. Sci.39(1999), 928-942, ChemDraw(TM) binary format, ChemDraw(TM) XML text format. Can write MDL Molfiles, CML, ChemDraw(TM) XML text format. Can also read and write any format supported by the current release of OpenBabel. Can export pictures in PNG, Windows bitmap (.bmp), Encapsulated PostScript (EPS), and Scalable Vector Graphics (SVG). Can generate 3-D structures with the help of the external program BUILD3D.
Online help, including tool tips. 13C-NMR prediction, based on Bremser W, Mag. Chem.23(4):271-275. 1H-NMR prediction, based on additive rules and functional group lookup methods, described in Pretsch, Clerc, Seibl, Simon, 'Tables of Spectral Data for Structure Determination of Organic Compounds', 2ed., 1989, Springer-Verlag. Simple IR prediction. Simple pKa estimation. Octanol-water partition coefficient estimation.
Reaction analysis: gas-phase enthalpy change estimate, 1H NMR and 13C NMR comparison. Windows 95/98/NT version (warning: outdated!). Integration with OpenBabel, allowing XDrawChem to read and write over 20 different chemical file formats.
Xdrawchem For Mac Pro
URL Versions.